Gradient descent (GD) is by far the most popular optimization strategy used in machine learning and deep learning at the moment. It is used when training data models, can be combined with every algorithm and is easy to understand and implement. Everyone working with machine learning should understand its concept. We’ll walk through how to implement the gradient descent algorithm in an artificial neuron pf our own.
Notes
1. Import libraries
- numpy for mathematical operations
- matplotlib for data visualization
- sklearn.datasets for work on it
- accuracy_score for evaluation
1
2
3
4
5
| import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from sklearn.metrics import accuracy_score
|
Personalize graph
1
2
3
4
5
| plt.style.use('dark_background')
plt.rcParams.update({
"figure.facecolor": (0.12 , 0.12, 0.12, 1),
"axes.facecolor": (0.12 , 0.12, 0.12, 1),
})
|
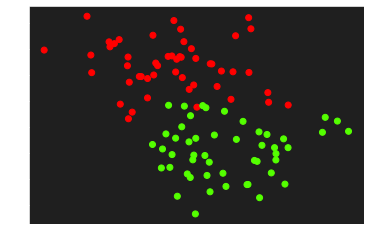
Let’s generate some data to practice with the make_blobs function from sklearn.dataset by specifying the number of inputs, the number of features, the centers and the random state
1
2
3
4
5
6
7
8
9
10
11
| # Generate data to practice
X, y = make_blobs(n_samples=100, n_features=2, centers=2, random_state=0)
# reshaping for a better adaptation to the calculation
y = y.reshape((y.shape[0], 1))
print('Dimension of X:', X.shape)
print('Dimension of y:', y.shape)
plt.scatter(X[:,0], X[:, 1], c=y, cmap='prism')
plt.show()
|
1
2
| Dimension of X: (100, 2)
Dimension of y: (100, 1)
|
2.Define Functions
Initialization
In order to create the initialization parameters of our separation line, i.e. different weights of the synaptic connections as well as the bias, the whole from the generated sample data
1
2
3
4
| def initialization(X):
W = np.random.randn(X.shape[1], 1)
b = np.random.randn(1)
return (W, b)
|
Model creation
To create the equations of the separation line and the activation function that will allow us to make better predictions. This function returns the active input
1
2
3
4
| def model(X, W, b):
Z = X.dot(W) + b
A = 1 / (1 + np.exp(-Z))
return A
|
Logarithmic Loss
In order to determine how close or far we are from the exact value after our prediction
1
2
| def log_loss(A, y):
return 1 / len(y) * np.sum(-y * np.log(A) - (1 - y) * np.log(1 - A))
|
Gradient
To apply more efficiently the equations related to gradient descent
1
2
3
4
| def gradients(A, X, y):
dW = 1 / len(y) * np.dot(X.T, A - y)
db = 1 / len(y) * np.sum(A - y)
return (dW, db)
|
Update Function
To apply the gradient descent
1
2
3
4
| def update(dW, db, W, b, learning_rate):
W = W - learning_rate * dW
b = b - learning_rate * db
return (W, b)
|
Predict Function
To predict the class of an input
1
2
3
4
| def predict(X, W, b):
A = model(X, W, b)
print(A)
return A >= 0.5
|
3. Create a Neuron
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
| def artificial_neuron(X, y, learning_rate = 0.1, n_iter = 150):
# Initialization W, b
W, b = initialization(X)
Loss = []
# Iteration loop
for i in range(n_iter):
A = model(X, W, b)
Loss.append(log_loss(A, y))
dW, db = gradients(A, X, y)
W, b = update(dW, db, W, b, learning_rate)
y_pred = predict(X, W, b)
print(accuracy_score(y, y_pred))
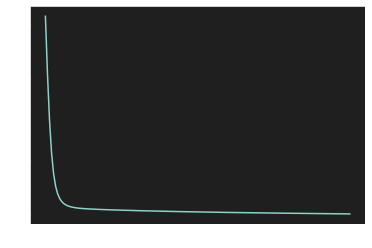
plt.plot(Loss)
plt.show()
return (W, b)
|
4.Visualization of the learning curve
1
| W, b = artificial_neuron(X, y)
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
| [[9.80406021e-01]
[7.67042239e-01]
[3.40212232e-03]
[1.12525858e-01]
[9.75128801e-01]
[3.45771392e-01]
[6.01324891e-02]
[9.67989464e-01]
[3.78178879e-02]
[8.76950867e-01]
[2.42449942e-02]
[8.88622356e-01]
[2.13762006e-02]
[1.24436424e-02]
[7.41091849e-01]
[9.90674953e-01]
[9.94228232e-01]
[2.71722474e-02]
[7.26832689e-01]
[6.53086468e-01]
[3.81186926e-02]
[2.84476759e-02]
[3.89230890e-01]
[3.35613428e-03]
[9.63415884e-01]
[2.57800801e-02]
[9.00335852e-01]
[6.60403491e-03]
[5.91310866e-02]
[7.80684024e-01]
[9.83746413e-01]
[3.66461272e-02]
[6.68889861e-01]
[9.84074985e-01]
[4.10732753e-01]
[2.41459372e-01]
[7.92174340e-01]
[5.77816615e-01]
[4.95755288e-01]
[4.29962960e-01]
[5.10137463e-02]
[7.64492353e-02]
[4.79641209e-04]
[1.56666125e-01]
[1.79877131e-01]
[8.65087917e-01]
[9.73102525e-01]
[9.66838846e-01]
[2.83194027e-03]
[6.89536120e-03]
[9.49692444e-01]
[5.48293144e-01]
[4.59304399e-02]
[3.52285969e-02]
[8.62738290e-01]
[3.57490916e-02]
[7.62844063e-01]
[8.40989205e-01]
[9.63965862e-01]
[9.93446898e-01]
[6.61394544e-01]
[1.82090079e-01]
[3.23128502e-03]
[9.44445420e-01]
[1.15292589e-02]
[3.68069696e-01]
[2.62313035e-02]
[6.74894046e-01]
[9.59434717e-01]
[2.76641732e-01]
[1.65157243e-01]
[9.29801123e-01]
[9.81566397e-01]
[1.06083626e-01]
[9.58192444e-02]
[2.87442858e-02]
[5.24151565e-01]
[8.97775539e-01]
[3.70343678e-02]
[2.80549476e-02]
[2.62000271e-01]
[8.94007894e-02]
[2.67674156e-03]
[1.11281924e-01]
[8.11706942e-02]
[8.72276677e-03]
[9.79280903e-01]
[6.77017113e-02]
[8.19492552e-01]
[9.67327252e-01]
[9.24699396e-01]
[9.86395708e-01]
[4.45440388e-01]
[9.33605778e-01]
[3.39032074e-01]
[6.16076544e-04]
[9.80401775e-01]
[9.73055392e-01]
[6.16480769e-03]
[4.11420426e-01]]
0.89
|
1
2
| [[ 1.42330917]
[-1.1221567 ]] [0.41562285]
|
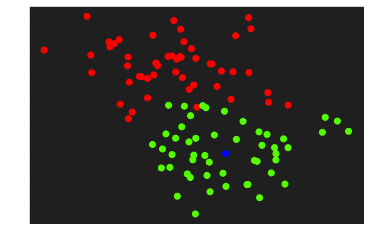
Let’s visualize and predict the class of a new element
1
2
3
4
5
6
7
| element = np.array([2,1])
plt.scatter(X[:,0], X[:, 1], c=y, cmap='prism')
plt.scatter(element[0], element[1], c='blue')
plt.show()
print(predict(element, W, b))
|
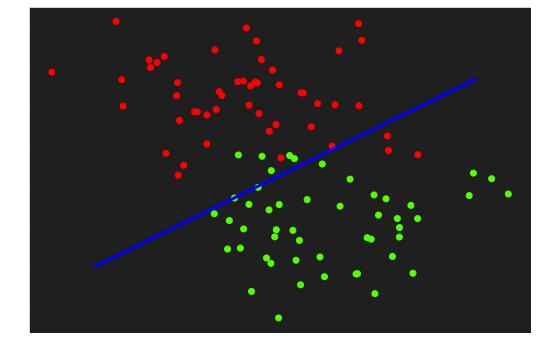
5. Descision border
Render the descision frontier
1
2
3
4
5
6
7
| fig, ax = plt.subplots(figsize=(9, 6))
ax.scatter(X[:,0], X[:, 1], c=y, cmap='prism')
x1 = np.linspace(-1, 4, 100)
x2 = ( - W[0] * x1 - b) / W[1]
ax.plot(x1, x2, c='blue', lw=3)
|
1
| [<matplotlib.lines.Line2D at 0x7f2c76337e80>]
|
6. 3D Visualization
1
| import plotly.graph_objects as go
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| fig = go.Figure(data=[go.Scatter3d(
x=X[:, 0].flatten(),
y=X[:, 1].flatten(),
z=y.flatten(),
mode='markers',
marker=dict(
size=5,
color=y.flatten(),
colorscale='YlGn',
opacity=0.8,
reversescale=True
)
)])
fig.update_layout(template= "plotly_dark", margin=dict(l=0, r=0, b=0, t=0))
fig.layout.scene.camera.projection.type = "orthographic"
fig.show()
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
| X0 = np.linspace(X[:, 0].min(), X[:, 0].max(), 100)
X1 = np.linspace(X[:, 1].min(), X[:, 1].max(), 100)
xx0, xx1 = np.meshgrid(X0, X1)
Z = W[0] * xx0 + W[1] * xx1 + b
A = 1 / (1 + np.exp(-Z))
fig = (go.Figure(data=[go.Surface(z=A, x=xx0, y=xx1, colorscale='YlGn', opacity = 0.7, reversescale=True)]))
fig.add_scatter3d(x=X[:, 0].flatten(), y=X[:, 1].flatten(), z=y.flatten(), mode='markers', marker=dict(size=5, color=y.flatten(), colorscale='YlGn', opacity = 0.9, reversescale=True))
fig.update_layout(template= "plotly_dark", margin=dict(l=0, r=0, b=0, t=0))
fig.layout.scene.camera.projection.type = "orthographic"
fig.show()
|
Links
Getting The Code On Google Colab
The entire code containing all the code mentioned in this post can be found here.
Getting The Code On Github
The entire folder containing all the code mentioned in this post can be found via this link.
Just bear in mind that you will need to install all the dependencies. If you find any issues with the code, feel free to either comment down below or raise an issue on Github.
Add me on LinkedIn
Don’t hesitate to follow me on linkedIn or o other social network to encourage me to do more posts on IT.
Comments powered by Venom Cocytus.